Cellular Genetics Informatics
Cellular Genetics
About us
We provide informatics support for the Human Cell Atlas, Human Induced Pluripotent Stem Cell Initiative and Human BioMolecular Atlas Program. We work with large amounts of single-cell RNA sequencing and microscopy imaging data.
More information about our group is available in our Travel Guide.
Our Services
Our team:
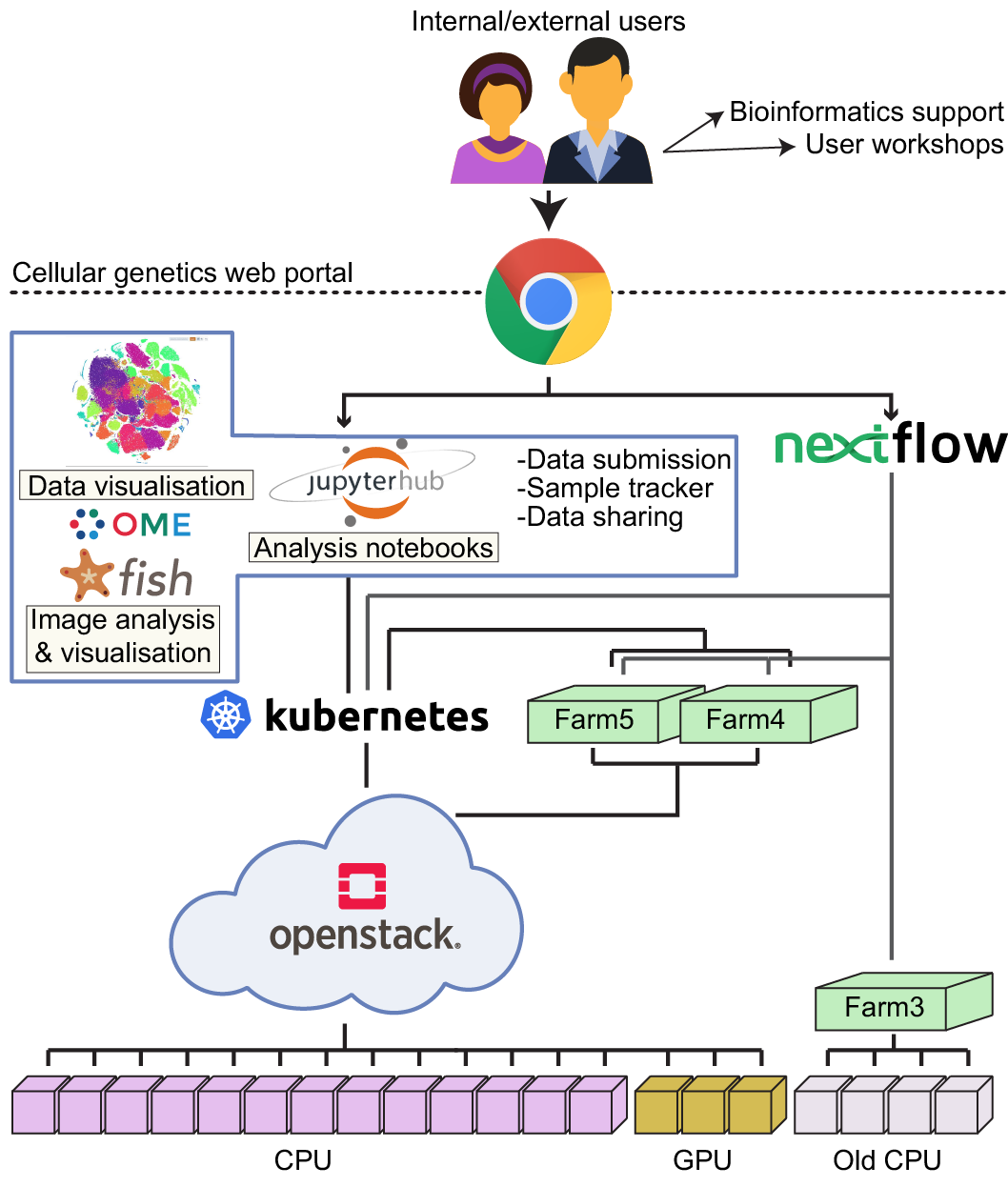
- provides comprehensive bioinformatics support for the Cellular Genetics programme
- develops and supports single-cell Nextflow analysis pipelines for various experimental protocols, including 10X, Smartseq2, scATACseq, TraCeR/BraCeR, CITEseq, inDrop. All of our pipelines have full HPC (LSF) and OpenStack Cloud integrations and are avaiable at GitHub
- enables data sharing with external collaborators
- facilitates data submission to ArrayExpress
- maintains and supports the programme’s Jupyter Hub dedicated for the data secondary analysis. We provide software support and a set of standard analysis notebooks, including Seurat, scanpy, single cell data integration notebooks
- enables data access for external collaborators via Jupyter Hub
- develops the programme’s internal Web portal, which includes a multiuser sample tracker, ability for the users to run our pipelines from the web and integration with our Jupyter Hub
- develops and supports the Cellular Genetics Imaging portal which allows the users to both run the imaging analysis pipelines and visualise their results
- develops GPU-accelerated imaging pipelines
- deploys bespoke and the publication supporting websites containing models and concise data visualisations
- organises workshops for the users on different topics, including Git, Jupyter Notebooks, Docker/Cloud, Rstudio/Shiny, Nextflow.
Illustrator: Christina Usher
Our Software/Analysis Stack
We run our pipelines and perform analysis on the Sanger’s High Performance Compute cluster (thousands of cores orchestrated by LSF) and the Sanger’s OpenStack Flexible Compute environment (private OpenStack cloud with thousands of cores orchestrated by Kubernetes). We use the following software/analysis stack (more information is in our GitHub organisation):
- Back End: Kubernetes, Docker, Singularity, Python
- Front End: Javascript, React
- Interactive Analysis Environments: Jupyter Hub
- Pipelines runners: Nextflow
- Secondary Analysis: R, python, C, bash
- Imaging: Omero, Bio-Formats, Fiji, cellprofiler, StarFish
Positions available
Our team is growing, please check the Sanger vacancies website for more information.
We have an excellent experience with student internships and apprenticeships. We welcome students with their own funding (with a possibility of topping it up) to work on both infrastructure and research projects.
Our people
Core team

Batuhan Cakir
Bioinformatician

Jessica Cox
Software Developer

Dr Pavel V Mazin
Senior Bioinformatician

Dr Alexander Predeus
Principal Bioinformatician

Martin Prete
Senior Software Developer
Previous core team members

Miss Tanya Bayzetinova
Student Placement

Batuhan Cakir
Visiting Scientist

Mr Ruben Chazarra Gil
Visiting Scientist

Dr Maria Keays
Senior Software Developer

Anton Khodak
Senior Software Developer

Dr Vladimir Kiselev
Cellular Genetics Informatics Team Leader

Simon Murray
Bioinformatician

Aleksandra (Ola) Tarkowska
Solutions Architect

Dr Stijn van Dongen
Senior Software Developer